Biography
Ryan R. Cheng is an assistant professor of chemistry at the University of Kentucky. His research focuses on using computer simulations, biopolymer theory, statistical mechanics, and machine learning to address biophysical problems, particularly in the area of structural genomics.
Interests
- Biophysics
- Computational and Theoretical Chemistry
Education
Postdoc/Research Scientist, 2022
Center for Theoretical Biological Physics, Rice University
Ph.D. in Theoretical Chemistry, 2012
University of Texas at Austin
B.S. in Chemistry, 2007
Carnegie Mellon University
The Team
*





Aleena Javaid, Undergraduate Researcher
Aleena is currently a 3rd year chemistry student at UKY.

Darin Vaughan, Graduate Student
Darin is a graduate student in chemistry and biomedical data scientist for the Innovation in Population Health Center at UK. His specialties are mathematical modeling of cell signaling and statistical linear algebra algorithms.

Devilal Dahal, Postdoctoral Researcher
Devilal received his Ph.D. in Computational Physics from the University of Southern Mississippi under the supervision of Professor Parthapratim Biswas. His research currently focuses on the mechanics of the genome.

Matheus Mello, Graduate Student
Matheus is currently a Ph.D. student at Rice University under the supervision of Professor Jose Onuchic. We are currently collaborating on the analysis of imaging data!

Sasindu Gunasinghe, Graduate Student
Sasindu received his bachelors degree in Chemistry from the Institute of Chemistry Ceylon in Sri Lanka. His research will focus on the role of epigenetics in cell differentiation and disease states.

Virangi Hewage, Graduate Student
Virangi is a Ph.D. student in chemistry who is interested in the conformational dynamics of proteins and protein-protein interactions.

Projects
*
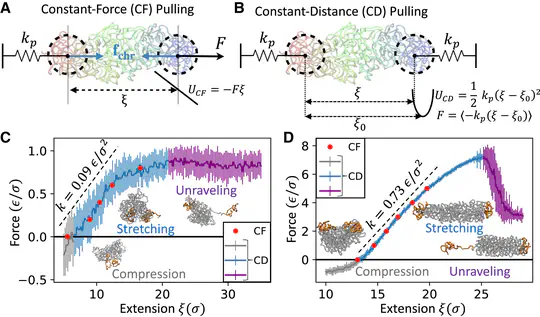
We are investigating how mechanical deformations modulate the structures and dynamics of the human genome.
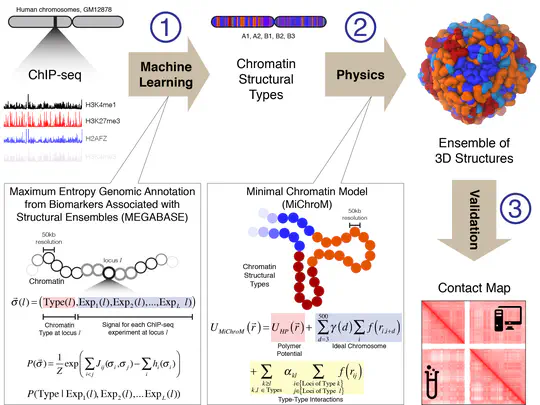
We explore the 3D structures of chromatin and the relationship of those structures to biological functions, such as transcriptional regulation.
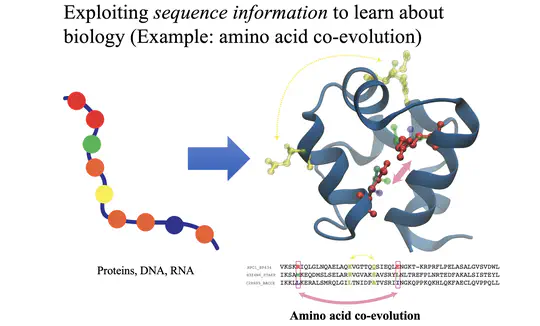
Amino acid coevolution is used to predict spatial contacts that have been maintained over the course of natural selection.
Publications
(2024).
Residue coevolution and mutational landscape for OmpR and NarL response regulator subfamilies.
Biophysical Journal.
(2023).
3D genome architecture regulates the traffic of transcription factors throughout human chromosomes.
Biophysical Journal.
(2023).
Structural Integrity and Relaxation Dynamics of Axially Stressed Chromosomes.
APS March Meeting Abstracts.
(2023).
Structural reorganization and relaxation dynamics of axially stressed chromosomes.
Biophysical Journal.
(2022).
Uncovering the statistical physics of 3D chromosomal organization using data-driven modeling.
Current Opinion in Structural Biology.
Recent & Upcoming Talks
Alumni
*

Justin Denny, Undergraduate Researcher
Justin graduated from UKY in 2023. He is currently a U.S. Army Officer & prospective medical student

Mayu Shibata, Graduate Student
Mayu received her Ph.D. from Ochanomizu University in March 2024 under the supervision of Professor Kei Yura. Her paper on amino acid co-evolution of bacterial response regulator proteins was published in Biophysical Journal in 2024. She is currently a researcher at the Center for Regenerative Medicine, National Research Institute for Child Health and Development (Japan).

Contact
Please feel free to email me!
- ryan.r.cheng@uky.edu
- (859)-257-3294
- 125 Chemistry/Physics Building, Office 351, Lexington, KY 40507