Protein sequence analysis
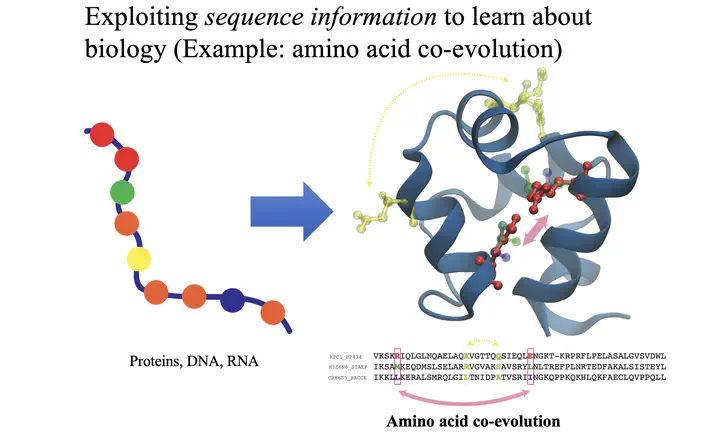
Statistical approaches such as Direct Coupling Analysis (DCA) (Morcos et al PNAS 2011) have revolutionized the prediction of 3D spatial contacts in proteins or protein complexes by using coevolutionary information extracted from protein sequence data. Coevolutionary information refers to the correlations between residues that reflect the maintainence of spatial contacts over the course of natural selection. We have previously worked extensively on quantifying the coevolution between residues of bacterial signaling proteins to maintain their signaling specificity (Cheng et al PNAS 2014, Cheng et al MBE 2016). We have also used DCA to predict the 3D structure of bacterial condensin (Krepel et al 2018), a motor protein complex that plays a key role in the organization of the bacterial genome. We are currently using probablistic modeling to infer a “nuclear interactome” of biomolecular interactions that play important roles in maintaining, regulating, and organizing the genome.